Release62:QMMM Free Energy
From NWChem
Contents |
QMMM Free Energy
Overview
Free energy capabilities of QM/MM module are at this point restricted to calculations of free energy differences between two fixed configurations of the QM region.
Users must be warned that this the least automated QM/MM functionality containing several calculation stages. Solid understanding of free energy calculations is required to achieve a meaningful calculation.
Description of the implemented methodology can be found in the following paper. In this approach the free energy difference between the two configurations of the QM region (e.g. A and B):
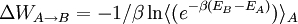
is approximated as a sum of internal QM contribution and solvation:
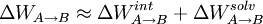
It is presumed that structures of A and B configurations are available as restart files sharing common topology file.
Internal contribution
The internal QM contribution is given by the differences in the internal QM energies evaluated at the optimized MM environment:
The internal QM energy is nothing more but a gas phase expression total energy but evaluated with the wavefunction obtained in the presence of the environment. To calculate internal QM contribution to free energy difference one has to
- Optimize MM environment for each configuration
- Perform total energy calculation for each configuration
- Calculate internal energy difference
Note that internal QM energy can be found in the QM/MM output file under "quantum energy internal" name.
Solvation contribution
The solvation contribution is evaluated by averaging energy difference between A and B configurations of the QM system represented by a set of ESP charges.
where is the total energy of the system where QM region is replaced by a set of fixed point ESP charges.
In majority of cases the A and B configuration are "too far apart" and one step free energy calculation as shown above will not lead to meaningful results. One solution is to introduce intermediate points that bridge A and B configurations by linear interpolation
where
The solvation free energy difference can be then written as sum of differences for the subintervals :
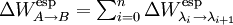
To expedite the calculation it is convenient to use a double wide sampling strategy where the free energy differences for the intervals and are calculated simultaneously by sampling around point. In the simplest case where we use two subintervals (n=2)
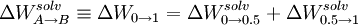
or
The following items are necessary:
- Restart file corresponding to either A or B configuration of the QM region (sharing the same topology file)
- ESP charges for QM region in .esp format for both configurations
- Coordinates for QM regions in .xyzi format for both configurations
Both .esp and .xyzi files would be typically obtained during the calculation of internal free energy (see above). ESP charges would be generated in the perm directory
during optimization of the MM region.
The xyzi is basically xyz structure file with an extra column that
allows to map coordinates of QM atoms to the overall system. The xyzi file can also be obtained as part of calculation of internal free energy by inserting
set qmmm:region_print .true.
anywhere in the input file during energy calculation. Both xyzi and esp files should be placed into the perm directory!!!
In the input file the restart file is specified in the MD block following the standard notation
md system < name of rst file without extension> ... end
while coordinates of QM region (xyzi files) and ESP charges (esp files) are set using the following directives (at the top level outside of any input blocks)
set qmmm:fep_geom xxx_A.xyzi xxx_B.xyzi set qmmm:fep_esp xxx_A.esp xxx_B.esp
The current interpolation interval for which free energy difference is calculated is defined as
set qmmm:fep_lambda lambda_i lambda_i+1
To enable double wide sampling use the following directive
set qmmm:fep_deriv .true.
If set, the above directive will perform both and ![\,\! [\lambda_i\to\lambda_{i-1}]](../../images/math/a/8/5/a85f5cafda04fbc6528367b9ed9ae67d.png) calculations, where
calculations, where
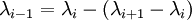
The calculation proceeds in cycles, each cycle consisting of two phases. First phase is generation of classical MD trajectory around point. Second phase is processing of the generated trajectory to calculate averages of relevant energy differences. The number of MD steps in the first phase is controlled by the QM/MM directive
- nsamples <integer number of MD steps for sampling>
This is a required directive for QM/MM free energy calculations.
Number of overall cycles is defined by the QM/MM directive
- ncycles <integer number of cycles default 1>
In most cases explicit definition of QM/MM density and region should not be required. The QM/MM density will automatically default to espfit and region to mm.
Prior to data collection for free energy calculations user may want to prequilibrate the system, which can be achieved by equil keyword in the MD block:
md ... equil <number of equilibration steps> end
Other parameters (e.g. temperature and pressure can be also set in the MD block.
The actual QM/MM solvation free energy calculation is invoked through the following task directive
task qmmm fep
The current value of solvation free energy differences may be tracked though
grep free <name of the output file>
The first number is a forward () free energy difference and second number is backward (![\,\! [\lambda_i\to\lambda_{i-1}]](../../images/math/a/8/5/a85f5cafda04fbc6528367b9ed9ae67d.png) ) free energy difference, both in kcal/mol. The same numbers can also be found in the 4th and 6th columns of <system>.thm file but this time in atomic units.
) free energy difference, both in kcal/mol. The same numbers can also be found in the 4th and 6th columns of <system>.thm file but this time in atomic units.
The same <system>.thm file can also be used to continue from the prior calculation. This will require the presence of
set qmmm:extend .true.
directive, the <system>.thm file, and the appropriate rst file.
Here is an example of the input file for QM/MM solvation free energy calculation.
